Relationship between Single Nucleotide Polymorphism of IGF-1 Gene in Nile Tilapia (Oreochromis niloticus) and Weight Gain

Abstract
Production and consumption of Nile tilapia (Oreochromis niloticus) have been growing. Insulin-like growth factor-1 (IGF-1) is a hormone that affects animal weight. Traditional breeding combined with molecular marker technology-assisted selective breeding has long been used to cultivate new fish strains and produce high-quality Nile tilapia. Moreover, molecular markers or genes related to important economic traits can be identified with the help of molecular-assisted selection breeding. A total of 152 Nile tilapia fish fin samples were collected from Janghang fish farm and classified according to their weight and sex. A total of 9 single nucleotide polymorphisms (SNPs) were identified in the Nile tilapia IGF-1 gene in this study. One SNP was located in the promoter region, three in the 5'untranslated regions (UTR), and five in the 3'untranslated regions (UTR). Effective genotypic blocks (GBs) were selected by analyzing the correlation between body weight and GBs. These molecular marker findings will facilitate the improvement of the Nile tilapia species in the future and enable the selection of fast-growing new strains, shorten the breeding cycle, achieve maximum economic benefits, and realize the important economic traits of tilapia.
Keywords:
Nile tilapia, Body weight, Principal component analysis, Insulin-like growth factors –1, Single nucleotide polymorphismⅠ. Introduction
Nile tilapia (Oreochromis niloticus) is a fish of the cichlid family. Insulin-like growth factor 1 (IGF-1) is a member of the insulin family and is an important gene involved in animal growth (Teng et al. 2020; Aljuboory and Alkhshali, 2018; Aljuboory and Alkhshali, 2018; Breidy Lizeth Cuevas-Rodríguez et al., 2016). IGF-1 regulates animal growth, development and metabolism, and plays an important role in regulating cell growth and differentiation. Researches of SNPs are also an important species improvement method in modern aquatic research (Liu et al., 2007; Feng, Yu, and Tong, 2014; Zhang et al., 2016). In addition, monitoring SNPs contributes to the traceability of many cases of aquatic fish diseases (Feng Ying et al., 2018; Nie et al., 2019; Gu et al., 2018). Variations in IGF 1 gene had an important effect on the muscle growth and body development of tilapia (Zeng et al., 2021; Laible, Wei, and Wagner, 2015). We will study the relationship between the IGF1 gene and body weight change during the growth of tilapia cultured in Janghang fish farm.
Ⅱ. Materials and Methods
1. Sample collection and extraction of gDNA
A total of 152 fish fin samples were collected from the batches. The Nile tilapia used in this experiment is a non-wild species collected at Janghang fish farm. This fishery has 12 cylindrical breeding tanks with a diameter of 6 meters and a water depth of 1.5 meters. The fry hatched from the same batch of fertilized eggs and was raised for 24 months. About 5,000-6,000 fish are put into each tank, The breeding water temperature of this batch of samples is controlled at 24-32°C, and the mature body whose growth cycle reaches six months is selected as the standard. To use the selected sample species for future breeding experiments, this experiment to investigates the relationship between SNPs and body weight. Their genomic DNA was extracted by DNeasy Blood & Tissue Kit (QIAGEN) from the collected fish fin samples.
2. PCR amplification and DNA sequencing analysis
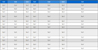
We designed tilapia-specific polymerase chain reaction (PCR) primers for the promoter region, 5'UTR, and 3'UTR of the IGF-1 gene, shown in table (<Table 1>) using the NCBI database sequence (IGF-1, GenBank accession number is NC_031981). DNA amplification was performed by PCR. The composition of the PCR master mix was as follows: 0.25 µl Ex Taq DNA polymerase, 5 µl Buffer 10x, 4 µl dNTPs, 2 µl Forward Primer, 2 µl Reverse Primer, 0.5 µl genomic DNA, and 36.25 µl of distilled water. The annealing temperature was calculated for each primer pair. The thermal cycle program was set to 94℃ for 5 minutes (pre-denaturation), 94℃ for 30 seconds for denaturation, annealing temperature for 30 seconds, and at 72℃ extension time depending on the length of the PCR product. A FavorPrep GEL/PCR Purification Mini kit was used for electrophoresis and gel purification of the PCR products, which were then submitted to Macrogen CO (Macrogen Building, 238 Teheran-ro, Gangnam-gu, Seoul, Korea).
3. Discovery of SNP and analysis the relation between the genotype block and body weight
For SNP detection, the BioEdit (7.0.9.0) alignment tool was used to align the sequences, followed by analysis with ABI sequencing technology. Clusters of SNP regions are divided into individual blocks using a parental genotyping assignment referred to as genotype blocks (GBs).
4. Statistical analysis
In order to investigate the link between the genotype block of the SNP and the body weight features of tilapia, one-way ANOVA was performed using SPSS Statistical software version 22.0. Data are provided as means ±SD, with a significance level of P≤0.05. By calculating the amount of distinct body weight characters in each SNP and utilizing the various genotype blocks of SNPs as factors and the body weight traits of tilapia as dependent variables, the differences in body weight traits across different genotype blocks were investigated.
5. Principal component analysis
Using the R programming language, PCA was performed and visualized in this study The available built-in packages were used to complete all calculations. Variables were coded for the various IGF-1 SNPs found in the S(k) datasets. A k range of 0.30 to 315.00 was given to the S(k) data set. To represent a single data point for each vanable in the PCA calculation, a certain set of S(k) values were chosen. Using Factoextra reference packages, the PCA findings were additionally visualized. The Factoextra package, which also showed a classification of the relevant factors, was used to create the loading graphs. Using the quality's eigenvalues, the quality of representations was assessed. With this package, such values are commonly represented by a color gradient and normalized to a range of -1 to 1.
Ⅲ. Results
1. Analysis and classification of sample DNA results
This study was helpful as we explored effective techniques for discovering SNPs in the IGF-1 gene that can regulate muscle growth traits. We found a total of 9 SNPs in the IGF-1 gene, of which one site (~80bp) was found at the promoter region with a C/T change, and 3 sites were found in the 5'UTR at ~63bp (G/C), ~404bp (G/A), and ~443 bp (G/C). In the 3'UTR, we found a total of five SNPs at 18255bp (T/A), 18280bp (G/A), 18381 (G/A), 18425 (C/T), and 18456 (G/T). The location, base types, and the percentages of each SNPs were shown in <Tables 2> and <Tables 3>.
The average value of polymorphism was higher in exon regions than in intron and promoter regions.
2. Summary of analysis sample results
The relationship between the Genotype distribution and average body weight of male Nile tilapia is described in detail in <Table 4>. There are a total of 23 genotypes, The average individual body weight ranges from 1.33 kg to 0.61 kg.
With a total of 18 genotypes, <Table 5> details the link between the genotype distribution and average body weight of female Nile tilapia. The average body weight of an individual might range from 1.18 kg to 0.30 kg.
The results were combined according to the above data analysis; samples with the same genotypes were combined, and repeated similar genes were deleted. As shown in Table 6, a total of 32 fish samples with different genotypes were obtained for reference, Each genotype block (GB) with specific SNP type were showm in Table 6. GBF1 to GBF9(GBF: Genotype Blocks IGF-1) are the common genotypic blocks of male and female samples. GBF10 to GBF23 are unique genotypes for male samples, GBF24 to GBF32 are unique genotypes for female samples, and the GBF9 genotype is the wild-type genotype.
3. Sample statistical analysis
The genotype of each male sample was considered separately, and the average value and the percentage of male samples overall was calculated according to the weight score. As shown in Figure 1, the following excellent genotypes were selected: GBF1, GBF2, GBF3, GBF4, GBF8, and GBF10.
![[Fig. 1] [Fig. 1]](/xml/36394/KSFME_2023_v35n2_222_f001.jpg)
The relationship between male tilapia genotype block and body weight.*The X-axis and Y-axis represents the genotype-categorized male samples and their weight. The dotted line represents the average value of male samples 1.02kg. Green is the coloring of high-quality samples. Red is the coloring of poor-quality samples. Black and white are samples that are close to the average.
Considering the genotypes of the female samples separately, the average was calculated by weight and data from half of the total female samples. As shown in Figure 2, the excellent genetic samples we identified were as follows: GBF1, GBF2, GBF3, GBF24, GBF25, GBF26, and GBF27.
![[Fig. 2] [Fig. 2]](/xml/36394/KSFME_2023_v35n2_222_f002.jpg)
The relationship between female tilapia genotype block and body weight. *The X-axis represents the female samples classified by genotype and the Y-axis weight. the dotted line represents the average weight of female samples 0.63kg. Green is to highlight high-quality samples, red is for inferior samples, and black and white are close to the average the sample is mediocre.
4. principal component analysis
A statistical method was used to transform a group of potentially correlated variables into a group of linearly uncorrelated variables through orthogonal transformation, and this group of variables after conversion is called the principal component. In actual subjects, many related variables (or factors) are often put forward to analyze the problem comprehensively because each variable reflects some information about the subject to varying degrees. When using statistical analysis methods to study multivariate topics, too many variables will increase the complexity. People naturally hope that fewer variables will result in more information. In many cases, there is a certain correlation between variables. When there is a certain correlation between two variables, the two variables reflect an overlap in information about the subject to a certain degree. A principal component analysis removes redundant variables (closely related variables) and establishes a smaller number of uncorrelated ones while retaining the original information as much as possible. This statistical method, the principal component analysis, is also used in mathematics to reduce dimensionality. It is created by collecting the body weight of the Nile tilapia the DNA sample was extracted from and determining its relationship with the average value.
As shown in the PKC diagram in Figure 3 and Figure 4, male GBF1 and female GBF2 is a specific genotypes with an outstanding phenotype.
![[Fig. 3] [Fig. 3]](/xml/36394/KSFME_2023_v35n2_222_f003.jpg)
PCA analysis of male tilapia samples.*The weight of Nile tilapia was measured and recorded, and the sample DNA was extracted to further determine the genotype and classification. Single nucleotide polymorphism is the measurement point of the genotype variable of each sample in this study. In this figure, the Y-axis represents dimension 1 (dim1), and the calculated percentage is 43.3%, while the X-axis represents dimension DIM2, and the calculated percentage is 43.3%.
![[Fig. 4] [Fig. 4]](/xml/36394/KSFME_2023_v35n2_222_f004.jpg)
PCA analysis of female tilapia samples.*The weight of Nile tilapia was measured and recorded, and the sample DNA was extracted to further determine the genotype and classification. Single nucleotide polymorphism is the measurement point of the genotype variable of each sample in this study. In this figure, the Y-axis represents dimension 1 (dim1), and the calculated percentage is 30.7%, while the X-axis represents dimension DIM2, and the calculated percentage is 30.7%.
Measure and record the body weight of Nile tilapia to extract sample DNA to further determine genotype and classification. Single nucleotide polymorphism is the measurement point of the genotypic variables of each sample in this study. In Figure 4, the Y-axis represents dimension 1 (Dim1), and the calculated percentage is 43.3%, while the X-axis represents the calculated percentage of dimension Dim2 is 43.3%. In Figure 5, the Y-axis represents dimension 1 (Dim1), and the calculated percentage is 30.7%, while the X-axis represents the dimension Dim2’s calculated percentage is 30.7%. The closer the Y-axis and X-axis percentages are, the more sufficient sample collection is and the more universal it is.
Ⅳ. Conclusion
According to the data of this study, it is concluded that IGF-1 plays a crucial regulatory role in the growth of Nile tilapia. According to the 9 controllable SNPs found in the IGF-1 gene, we can select individuals with excellent genotypes and after special cultivation, obtain high-quality biological products and food for humans. The use of SNP screening is an efficient and rapid method of species improvement experimentation.
V. Discussion
Nile tilapia (Oreochromis niloticus) has an increasing significance in aquaculture worldwide (Kuebutornye et al., 2020; Marjanovic et al., 2016; Zou et al., 2015; Kishawy et al., 2020; Richter et al. 2021; Wardani et al., 2021). Other applications of QTL technology in aquatic research have also produced extensive and far-reaching significance (Fernández, Villanueva, and Toro, 2021; Mohammed, Raji, and Igwebuike, 2020; Sullivan et al., 2015). In this study, we used individual samples from Nile tilapia to explore the relationship between SNPs in the IGF-1 gene and fish growth. Other contemporary research studies on IGF-1 in other animals helped our experiments (Lu et al., 2020; Tsai et al., 2014; Wuertz et al., 2006). We found a total of 9 SNPs. The body weight of male fish was commonly greater than that of female fish. The difference in growth and the the average growth rate varies with gender. We classified individual fish according to gender and weight. Due to the increasing demand for Nile tilapia genetic research, food sources, and aquaculture industry, many countries have focused on cultivating Nile tilapia, enriching human food nutrition sources, and high-quality biological commodity breeding industry and quality improvement projects for other species (Zhiyun et al., 2016; Zhou et al., 2019). The study of genetic markers provides key information and helps select individuals with excellent genotypes, conduct experiments, explore optimum breeding options, and improve species improvement (Agarwal et al., 2014). This article demonstrates the connection between IGF-1 SNPs and tilapia growth. These collected data demonstrate the importance of genetic research and exploring the regulatory roles of other fish growth genes. Nile tilapia performed well for marker-assisted sample selection.
Acknowledgments
This research was funded by the Development of software for data collection and analysis of growth rate according to environment factor of (semi) circulation filtration system (20180352).
References
- Aljuboory ST and Alkhshali MS(2018). The Relation of Insulin like Growth Factor Gene (IGF-1) with Some Physiological Characteristics of Common CARP (Cyprinus Carpio). Journal of Entomology and Zoology Studies 6(1), 1300~1303. https://www.researchgate.net/publication/333486251
- Aljuboory ST and Alkhshali MS(2018). Relation of Insulin like Growth Factor Gene (IGF- 1) WHTH Chemical Analysis Common Carp (Cyprinus Carpio). Journal of Entomology and Zoology Studies 6(2), 1013~1016. https://www.researchgate.net/publication/333390256
-
Cuevas-Rodríguez BL, Sifuentes-RincónAna AM, Ambriz-Morales P, García-Ulloa M, Valdez-González FJ and Hervey Rodríguez-González(2016). Novel Single Nucleotide Polymorphisms in Candidate Genes for Growth in Tilapia (Oreochromis Niloticus). Revista Brasileira de Zootecnia 45(6), 345~48.
[https://doi.org/10.1590/S1806-92902016000600009]

-
Fernández J, Villanueva B and Toro MA(2021). Optimum Mating Designs for Exploiting Dominance in Genomic Selection Schemes for Aquaculture Species. Genetics Selection Evolution 53(1), 1~13.
[https://doi.org/10.1186/s12711-021-00610-9]

-
Gao FY, Lu MX, Cao JM, Liu ZG, Zhu HP, Ke XL and Yang X Le(2018). Screening of IPS - 1 Gene SNPs and Their Association with Resistance to the Infection of Streptococcus Agalactiae in Oreochromis Niloticus. Journal of Agricultural Biotechnology 26(1), 20~33.
[https://doi.org/10.3969/j.issn.1674-7968.2018.01.003]

-
Gu XH, Jiang DL, Huang Y, Li BJ, Chen CH, Lin HR and Xia JH(2018). Identifying a Major QTL Associated with Salinity Tolerance in Nile Tilapia Using QTL-Seq. Marine Biotechnology 20(1), 98~107.
[https://doi.org/10.1007/s10126-017-9790-4]

-
Kuebutornye FKA, Abarike ED, Sakyi ME, Lu YS, and Wang ZW(2020). Modulation of Nutrient Utilization, Growth, and Immunity of Nile Tilapia, Oreochromis Niloticus: The Role of Probiotics. Aquaculture International 28(1), 277~91.
[https://doi.org/10.1007/s10499-019-00463-6]

-
Kishawy Asmaa-TY, Roushdy EM, Hassan Fardos-AM, Mohammed Haiam-A and Abdelhakim Taghrid-MN (2020). Comparing the Effect of Diet Supplementation with Different Zinc Sources and Levels on Growth Performance, Immune Response and Antioxidant Activity of Tilapia, Oreochromis Niloticus. Aquaculture Nutrition 26(6), 1926~1942.
[https://doi.org/10.1111/anu.13135]

-
Liu HN, Li S, Wang ZF, Hou P, He QG and He NY(2007). PCR Amplification on Magnetic Nanoparticles: Application for High-Throughput Single Nucleotide Polymorphism Genotyping. Biotechnology Journal 2, 508~11.
[https://doi.org/10.1002/biot.200600214]

-
Laible G, Wei JW and Stefan W(2015). Improving Livestock for Agriculture - Technological Progress from Random Transgenesis to Precision Genome Editing Heralds a New Era. Biotechnology Journal 10, 109~20.
[https://doi.org/10.1002/biot.201400193]

-
Lu S, Liu Yang, Yu XJ, Li YZ, Yang YM, Wei Min, Zhou Q., et al.(2020). Prediction of Genomic Breeding Values Based on Pre-Selected SNPs Using SsGBLUP, WssGBLUP and BayesB for Edwardsiellosis Resistance in Japanese Flounder. Genetics Selection Evolution 52(1), 1~10.
[https://doi.org/10.1186/s12711-020-00566-2]

-
Miaomiao N, Hu JW, Lu YL, Wu ZH, Wang LJ, Xu DD, Zhang PJ and You F(2019). Cold Effect Analysis and Screening of SNPs Associated with Cold-Tolerance in the Olive Flounder Paralichthys Olivaceus. Journal of Applied Ichthyology 35(4), 924~32.
[https://doi.org/10.1111/jai.13908]

-
Marjanovic J, Mulder HA, Khaw HL and Bijma P(2016). Genetic Parameters for Uniformity of Harvest Weight and Body Size Traits in the GIFT Strain of Nile Tilapia. Genetics Selection Evolution 48(1), 1~10.
[https://doi.org/10.1186/s12711-016-0218-9]

-
Mohammed A, Raji AO and Igwebuike J(2020). Genetic Variability of Nigerian Sheep Breeds at the Insulin-like Growth Factor-1 (IGF-1) Gene Locus. Nigerian Journal of Animal Production 45 (December), 51~60.
[https://doi.org/10.51791/njap.v45i1.367]

-
Pinky A, Parida SK, Mahto A, Das S, Mathew IE, Malik N and Tyagi AK(2014). Expanding Frontiers in Plant Transcriptomics in Aid of Functional Genomics and Molecular Breeding. Biotechnology Journal 9, 1480~92.
[https://doi.org/10.1002/biot.201400063]

-
Richter BL, Silva T.S-de.C, Michelato M, Marinho MT, Gonçalves GS and Furuya WM(2021). Combination of Lysine and Histidine Improves Growth Performance, Expression of Muscle Growth-Related Genes and Fillet Quality of Grow-out Nile Tilapia. Aquaculture Nutrition 27(2), 568~80.
[https://doi.org/10.1111/anu.13207]

-
Sullivan CV, Chapman RW, Reading BJ and Anderson PE(2015). Transcriptomics of MRNA and Egg Quality in Farmed Fish: Some Recent Developments and Future Directions. General and Comparative Endocrinology 221, 23~30.
[https://doi.org/10.1016/j.ygcen.2015.02.012]

-
Teng T, Zhao XQ, Li CJ, Guo JQ, Wang YF, Pan CL, Liu EG and L QF(2020). Cloning and Expression of IGF-I, IGF-II, and GHR Genes and the Role of Their Single-Nucleotide Polymorphisms in the Growth of Pikeperch (Sander Lucioperca). Aquaculture International 28(4), 1547~61.
[https://doi.org/10.1007/s10499-020-00542-z]

-
Tsai HY, Hamilton A, Guy DR and Houston RD (2014). Single Nucleotide Polymorphisms in the Insulin-like Growth Factor 1 (IGF1) Gene Are Associated with Growth-Related Traits in Farmed Atlantic Salmon. Animal Genetics 45(5), 709~15.
[https://doi.org/10.1111/age.12202]

- Tao ZY, Zhu CH, Song WT, Xu WJ, Liu HX, Zhang SJ and Li HF(2016). Polymorphism of IGF-1 Gene and Relationship between Genotypes and Skeletal Muscle Development Traits in Gaoyou Duck. Journal of Northeast Agricultural University 47, 89~92. http://www.cnki.net/kcms/detail/23.1391.S.20161228.1033.018.html
-
Wuertz S, Nitsche A, Gessner J, Kirschbaum F and Kloas W(2006). IGF-I and Its Role in Maturing Gonads of Female Sterlet, Acipenser Ruthenus Linnaeus, 1758. Journal of Applied Ichthyology 22 (SUPPL. 1), 346~52.
[https://doi.org/10.1111/j.1439-0426.2007.00982.x]

-
Wardani WW, Alimuddin A, Junior MZ, Setiawati M, Nuryati S and Suprayudi MA(2021). Growth Performance, Robustness against Stress, Serum Insulin, IGF-1 and GLUT4 Gene Expression of Red Tilapia (Oreochromis Sp.) Fed Diet Containing Graded Levels of Creatine.” Aquaculture Nutrition 27(1), 274~86.
[https://doi.org/10.1111/anu.13184]

-
Xiu F, Yu XM and Tong JG(2014). Novel Single Nucleotide Polymorphisms of the Insulin-Like Growth Factor-I Gene and Their Associations with Growth Traits in Common Carp (Cyprinus Carpio L.). International Journal of Molecular Sciences.
[https://doi.org/10.3390/ijms151222471]

-
Zeng D, Xiong G, Wang XQ, Zhou XW, Wang P, Peng N, Luo ZJ and Li X(2021). Transcriptome Analysis and Identification of Genes and Single-Nucleotide Polymorphisms Associated with Growth Traits in the Chinese Soft-Shelled Turtle, Pelodiscus Sinensis. Journal of the World Aquaculture Society 52(4), 913~31.
[https://doi.org/10.1111/jwas.12780]

-
Zou GW, Zhu YY, Liang HW and Li Z(2015). Association of Pituitary Adenylate Cyclase-Activating Polypeptide and Myogenic Factor 6 Genes with Growth Traits in Nile Tilapia (Oreochromis Niloticus). Aquaculture International 23(5), 1217~25.
[https://doi.org/10.1007/s10499-015-9878-7]

-
Zhang SY, Chen XH, Wang MH, Zhong LQ, Qin Q and Bian WJ(2016). Development and Characterization of Ten Polymorphic SNP Markers in Growth-Related Genes for the Channel Catfish, Ictalurus Punctatus (Rafinesque, 1818). Journal of Applied Ichthyology 32(6), 1248~51.
[https://doi.org/10.1111/jai.13194]

-
Zhou Y, Zhang XJ, Xu Q, Yan JP, Yu F, Wang FH, Xiao J, Luo YJ and Zhong H(2019). Nonadditive and Allele-Specific Expression of Insulin-like Growth Factor 1 in Nile Tilapia (Oreochromis Niloticus, ♀) × Blue Tilapia (O. Aureus, ♂) Hybrids. Comparative Biochemistry and Physiology Part - B: Biochemistry and Molecular Biology 232, 93~100.
[https://doi.org/10.1016/j.cbpb.2019.03.002]